| MetaBakeryCustomized BioBakery wmgx workflow | |
| |
|
 |
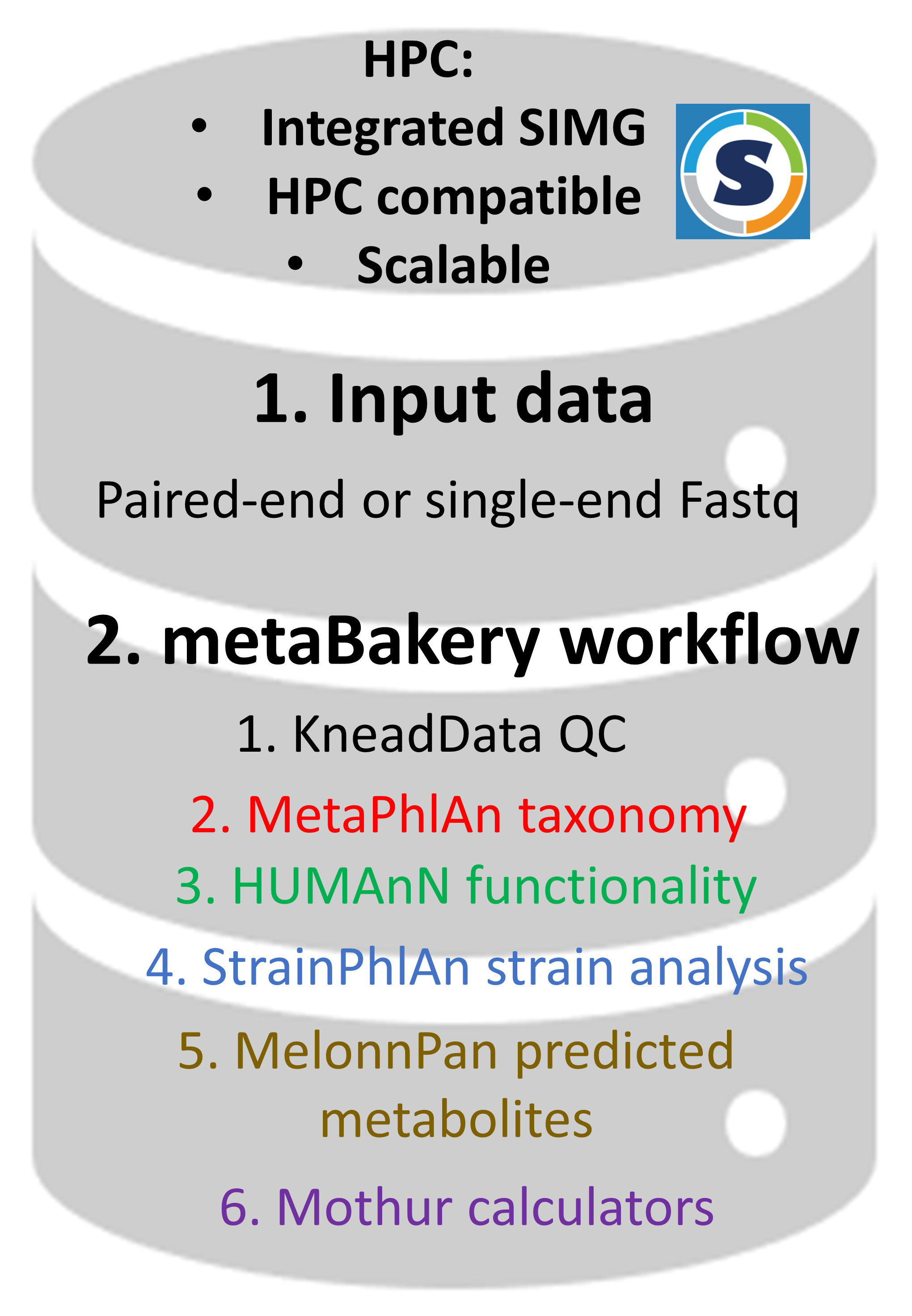
• all integrated programs and databases are fully preconfigured |
| • external databases may be used instead of the built-in ones | |
| • efficient utilization of computing resources | |
| • suitable for autonomous and batch execution | |
| • suitable for High-Performance Computing facilities | |
| • automatic crash recovery | |
| • possibility of splitting large datasets into manageable chunks and processing them separately (on different computers and/or HPC systems) | |
| • transparent handling of paired and unpaired reads (possibly intermixed) | |
| • transparent handling of major compression formats (gz, zip, bz2, xz), possibly intermixed | |
| • automatic handling of command-line parameters for included programs | |
| • experienced users can prescribe custom parameters | |
| • efficient restarts with changed parameters and input sets | |
| • complete screen and configuration dumping for easy documentation | |
| • easy access to command lines, exit codes and messages of programs | |
| • versions V4, V3 and V2 of BioBakery programs | |
| • only meaningful output files are presented to a user | |
| Licensing: Creative Commons Attribution CC BY licence |
|
IMPORTANT.
MetaBakery is a skeleton application for a synergic
execution of many externally developed pieces of software.
These are disseminated as integrated parts of MetaBakery to
provide the level of end-user experience that the system aims
to deliver. Nonetheless, every included piece of software
remains owned and copyrighted by its respective developers.
Please see Chapter Credits in MetaBakery Users' Manual for a list of included software. |
| IMPORTANT. MetaBakery is developed and disseminated in a good faith and desire to work according to expectations, but authors DO NOT give any guarantees about its correctness. USE IT AT YOUR OWN RISK. Authors cannot be held legally or morally responsible for any consequences that may arise from using or misusing MetaBakery. |
| Reference for citation |
| If you use MetaBakery, please cite the following reference. |
|
B. Murovec, L. Deutsch, D. Osredkar, B. Stres,
MetaBakery: a Singularity implementation of bioBakery tools as a skeleton application for efficient HPC deconvolution of microbiome metagenomic sequencing data to machine learning ready information, Frontiers in Microbiology 15/2025, https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2024.1426465, DOI=10.3389/fmicb.2024.1426465 |
|
Please also cite the included software that you use as part of your pipeline processing. Please see Chapter Credits in MetaBakery Users' Manual for a list of included software. |
| Download links |
| MetaBakery is available as a Singularity image, which may be run under Singularity 3.x. |
| Please consult MetaBakery Users' Manual for instructions and detailed descritions of below available editions. |
| The most recent MetaBakery version 2b was released on 19. 3. 2023. |
| Download link for MetaBakery 2b Edition BioBakery V4 (72 GB). |
| Download link for MetaBakery 2b Edition BioBakery V3 (63 GB). |
| Download link for MetaBakery 2b Edition BioBakery V2 (30 GB). |
| Please note. MetaBakery images are fairly large because they contain all needed and fully preconfigured programs and databases. |
| If large sizes make file transfers difficult, it is possible to download MetaBakery in smaller pieces. |
| Optionally, advanced users may want to check integrity of downloaded MetaBakery images by examining their SHA256 hashes, which are calculated with the sha256sum utility. In short: put MetaBakery downloaded images into a subdirectory together with file mbakery_2b_sha256.txt, and execute command sha256sum -c mbakery_2b_sha256.txt from this directory. Please note that only the actually present pieces are tested. For the rest, the command outputs "FAILED open or read", which is okay. |
| To quickly start using MetaBakery, please follow instruction in the Quick Start Guide. |
| MetaBakery Workflow is fairly customizable by means of configuration files. A good starting point for preparing a custom configuration file is the following configuration template with a fair amount of comments and explanations. |
| Download link for MetaBakery Configuration file template. |
| Contact | |
| We look forward to receive your feedback regarding the use of MetaBakery, potentially discovered bugs and issues, as well as comments and suggestions for further development of the system. | |
|
Blaž Stres (metagenomic related topics)
<mailto:blaz.stres@bf.uni-lj.si> University of Ljubljana, Biotechnical faculty, Slovenia University of Ljubljana, Faculty of Medicine, Slovenia UNI-LJ, Faculty of Civil and Geodetic Engineering, Slovenia Institute of Microbiology, University of Innsbruck, Austria |
Boštjan Murovec (computing related topics)
<mailto:bostjan.murovec@fe.uni-lj.si> University of Ljubljana, Faculty of electrical engineering, Slovenia Leon Deutsch <mailto:leon.deutsch@bf.uni-lj.si> University of Ljubljana, Biotechnical faculty, Slovenia |